
Department of Molecular Bioanalytics and Bioelectronics, Fraunhofer IBMT, Arthur-Scheunert-Allee 114-116, D-14558 Nuthetal, Germany
e-mail: nickisch@ibmt.fhg.de
 |
Markus von Nickisch-Rosenegk and Frank F. Bier
Department of Molecular Bioanalytics and Bioelectronics, Fraunhofer IBMT, Arthur-Scheunert-Allee 114-116, D-14558 Nuthetal, Germany e-mail: nickisch@ibmt.fhg.de |
 |
In bionanotechnology nanostructures based on DNA have been constructed.
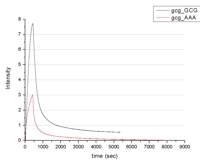
Generally it is a prerequisite to generate single stranded DNA (ssDNA) molecules to open the possibility of probing with oligonucleotides. The production of long ssDNA, which is suitable to address a sufficient amount of short sites with oligonucleotides, is not an easy task. Our experiments describe an alternative method to bind to dsDNA specifically and with a high affinity i. e. zinc finger (ZF) molecules. The aim is to develop zinc finger probes that are able to ligate at arbitrary sites of dsDNA molecules.
Two different zinc finger probes were commercially synthezised and modified with a Dy-633 fluorophore (Dyomics, Jena and PE GmbH, Nuthetal). The SP1 peptides were folded into functional zinc fingers using zinc chloride.
The addressable dsDNA was immobilized either on a biochip surface or on an optical fibre, where the kinetics and binding rates of the artifical zinc finger probes are measured by fluorescence detecting devices (chip-scanner, photomultiplying tube).
The sucessful experiments show a possible way to create dsDNA-binding probes by variation of synthezised zinc finger binding domains of the diverse existing natural zinc fingers. This might raise the chance to address dsDNA used in a nanoarray format.