Biomolecular Computing Devices
 |
Ehud Keinan
Department of Chemistry and Institute of Catalysis Science and Technology, Technion - Israel Institute of Technology, Technion City, Haifa 32000, Israel, and
Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, U.S.A.
e-mail: keinan@tx.technion.ac.il
URL: http://www.technion.ac.il/technion/chemistry/staff/keinan/
|
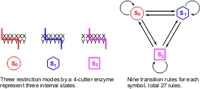 |
The first nanoscale, programmable 2-state-2-symbol finite automaton that computed autonomously with all of its components, including hardware, software, input and output being biomolecules, mixed together in solution was previously presented [1]. The hardware consisted of a restriction nuclease and a ligase, while the software (transition rules) and the input were double-stranded DNA oligomers. Computation was carried out by processing the input molecule via repetitive cycles of restriction, hybridization, and ligation reactions to produce a final-state output in the form of dsDNA molecules, which were characterized by gel electrophoresis. We increased the levels of complexity and mathematical power of these automata by the design of a 3-state-3-symbol automaton that has as many as 27 possible transition rules and a remarkable number of 939,524,089 syntactically distinct programs. This number is significantly larger than the corresponding number of 765 available programs with the 2-symbol-2-state device. Restrictions at the beginning of the symbol domain, 1 basepair deeper, and 2 basepairs deeper into the domain represent the three internal states, S0, S1 and S2, respectively, as shown below. The applicability of this design was further amplified by employing surface-anchored input molecules, using the surface plasmon resonance (SPR) technology to monitor the computation steps in real time. Computation was performed by alternating the feed solutions between BbvI endonuclease and a solution containing T4 DNA ligase, ATP and appropriate transition molecules. The output detection involved final ligation with one of three soluble detection molecules. Parallel computation and real-time detection were carried out automatically with a Biacore chip that carried 4 different inputs.



