Department of Chemistry and Biochemistry, University of California Santa Barbara, CA 93106-9510, U.S.A.
e-mail: achworos@chem.ucsb.edu
| Arkadiusz Chworos and Luc Jaeger
Department of Chemistry and Biochemistry, University of California Santa Barbara, CA 93106-9510, U.S.A. e-mail: achworos@chem.ucsb.edu |
 |
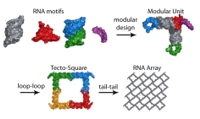
Nucleic acids are a highly predictable biosubstance and have been used previously to generate programmable architectures. Various geometrical objects like cube, octahedron or 2D lattices have been generated using multi cross-over of DNA molecules. Situated between DNA and encoded proteins in the biological pathway, ribonucleic acid provides an endless possibility 3D shapes as well as accurate prediction of the structures’ formation. Understanding of the assembly and folding principles of natural RNA led to build potentially useful artificial structures at the nano-scale level.
We design and build the RNA tecto-square by programming sequences using specific set of nucleotides that correspond to structural motifs. Each square is composed of four different RNA molecules joined together through specific loop-loop interactions. These tecto-squares can interact with each other through complementary dangling 3’-tails. A large variety of different, predictable 2D lattices have been generated by design of sequences combined with organization of loop and tail interactions. Atomic Force Microscopy technique has been successfully used for visualization of newly synthesized biolayers. Such programmable arrays can serve as a scaffold for deposition of nano-particles or quantum dots. Our system can further be used for the study of self-organization and/or self-assembly of nucleic acids on the surface.